What is biohacking?
(Art)ScienceBLR and Hackteria members were invited to the Swiss Citizen Science Network’s meeting “Biohacking meets Citizen Science” in Bern at Dialog Science and Society (Stiftung Science et Cité).
In the style of hackteria workshops, we started the morning hands-on.We set up the room as a temporary/mobile open source DIY bio-lab. During the round of self-introductions, we assembled gel-trays for electrophoresis. People were mainly from the German speaking parts of Switzerland, and Geneva’s Bioscope was represented – with educators, artists, biotechnologists, geologist, botanist…history of science – it was a nice atmosphere.
To use some of the generic lab equipment, we decided to extract some DNA from the participants and run a genomic PCR on the serotonin transporter. This was inspired by some protocols from the Science Bar Incubator in Tokyo.
Also, genotyping ourselves is a great starting point for a range of discussions on gene <> phenotype (can we tell something about a person’s personality just from differences in one gene?), genetic privacy, other techniques used…
Just to keep it simple, we decided to run one set of PCRs on the serotonin transporter (at the Science Bar, they run both the dopamine receptor D4R and the serotonin receptor) to look for their polymorphisms (different forms). The transporter recycles the serotonin released in between neurons back to the pre-synaptic neuron (the top neuron in the picture). Neurons communicate their electrical signal to the next neuron using neurotransmitters (small chemicals), such as dopamine and serotonin. Serotonin, amongst its many roles, is implicated in depression, and inhibition of the transporter is used to treat depression – and the transporter polymorphisms may be one of the factors affecting mood.
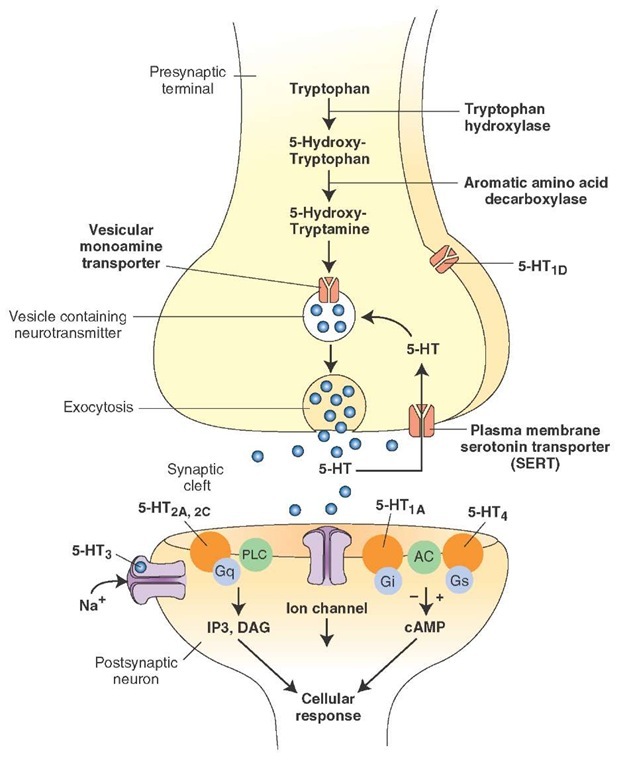
Serotonin in the synaptic cleft from http://what-when-how.com/
More on serotonin transporter (and from where this image was taken) can be found here.
We can do a simple PCR to look for the differences for this serotonin transporter, as there is a deletion, which results in the “Short” form (allele), which decreases the efficiency with which the serotonin transporter gene can be made (transcribed) into mRNA, and with less mRNA, less serotonin transporter protein, and less re-uptake of the serotonin between the neurons. We looked for the Long and the Short forms of the serotonin transporter gene.
The workflow was to
-collect some cells from the mouth
-extract the DNA using a detergent and a heat-activated protein degrading enzyme proteinase K (from a kit)
-take a small amount of the extracted DNA and use it as a template in a PCR reaction
-run an agarose gel electrophoresis to separate the long and short forms of the serotonin transporter gene
-to image the gel and make some conclusions
To collect the DNA, we took swabs of cells from our mouth.
Marc quickly made us a coconut sample holder as well 🙂
In parallel, we poured our gels and went through the rest of the work flow guided by Sachiko.
We used the home-built open PCR machine to extract the DNA and activate the proteinase K.
The primer sequences are from Gelernter et al. (1999).
-Forward primer: 5’- ATG CCA GCA CCT AAC CCC TAA TGT – 3’, (24)
-Reverse primer: 5’- GGA CCG CAA GGT GGG CGG GA – 3’. (20)
With 35 cycles at 98C (30′), 68C(30′), 72C(60′), the PCR products yield two identifiable forms – the Long form of 419 base pairs (bp), and the Short form of 376 bp.
We ran the gels in our DIY electrophoresis chamber and the voltage regulator.
Urs made this super neat gel visualiser, which we did not get to use, as we ran out of time.
We ran the gels again the next day at the Hackuarium where we got this result from an old machine with thermal paper printouts.
The gel we had poured did not contain the salts (so nothing to carry the current), so the gel running took a bit of time, and the bands are not as nicely separated as we would expect. This mistake is quite common in the lab. Our previous test runs looked more like this, where the long and short forms were more evident. The primers and PCR conditions are not absolutely perfect, as we saw many longer bands.
We also brought other prototypes to show – including the DIY fluorometer we will make at the Winter School in the next days.
Marc showed us his super microscopes, euglena and some bioluminescent algae.
This was followed by presentations from Marc, Shreyasi, Urs and Sachiko. Shreyasi introduced (Art)ScienceBLR.
The discussion following the presentations in the afternoon was extremely interesting.
We received some questions – where can we source all of these materials? and critical feedback – whether biohacking could be the bourgeois “gentleman science” of the last century, as most of us had some training and came from a standing of societal privilege. We also spoke about where exactly do the citizens come in and what role can/do they play in “citizen science”. We are citizens. You are citizens. Who are the citizens in citizen science???
The only thing that was missing was the field trip, which is usually an integral part of a hackteria workshop.